Exploring the universal healthy human gut microbiota around the World
In regards to the importance of better characterizing the gut microbiota, Piquer-Esteban et al. developed an enrichment strategy that improves the classification capacity compared to the original NT database.
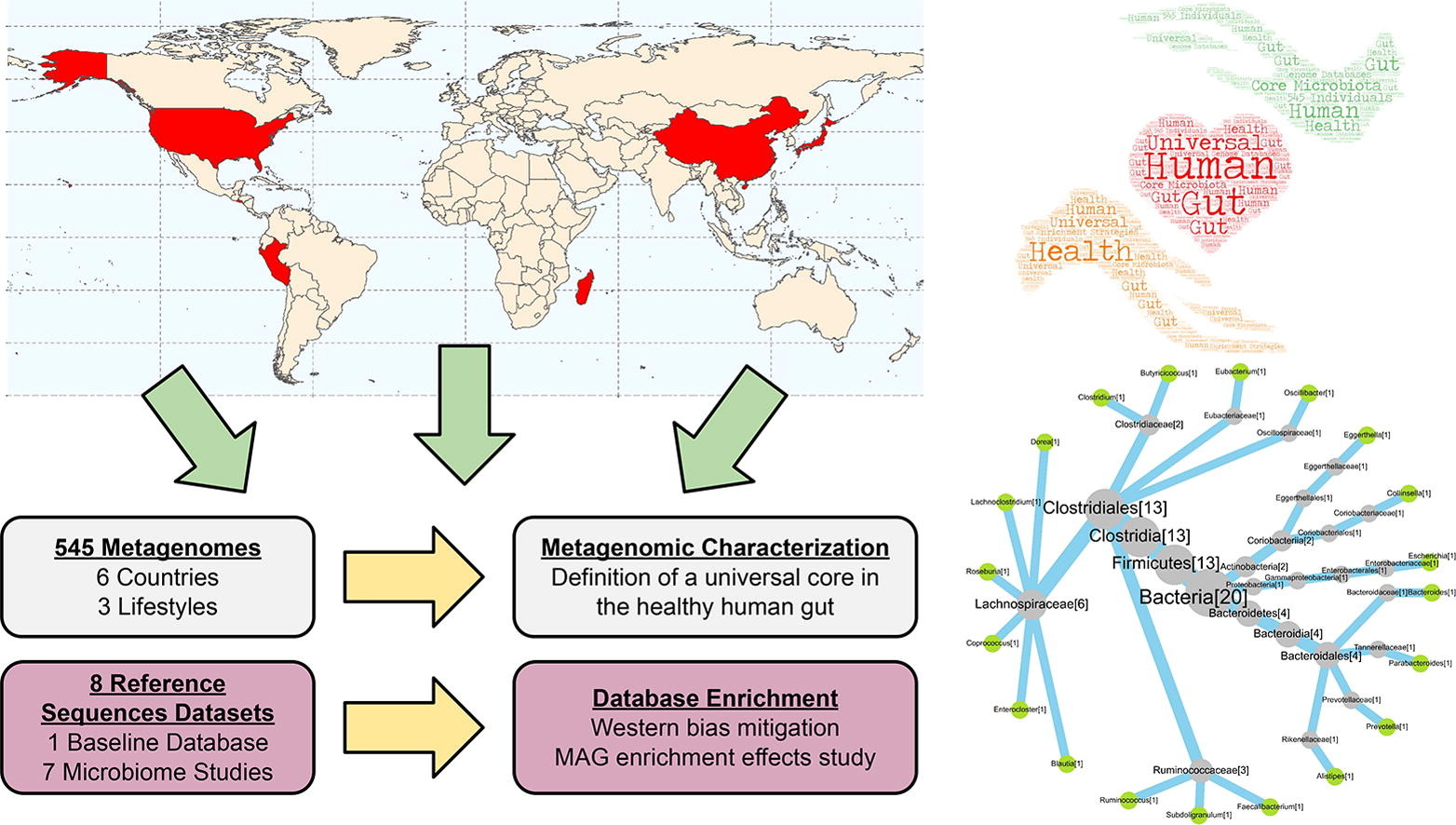
The human gut is extensively studied in terms of its microbial environments due to evidential link between the gut microbiota and host health. Yet, there is still a lack of knowledge about the core taxa forming the gut microbiota and, moreover, available information is biased towards western microbiomes in both genome databases and most core taxa studies.
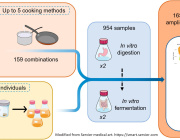
This study tackled those limitations by testing a database enrichment strategy and analyzing public datasets of whole-genome shotgun data, generated from 545 fecal samples, comprising three gradients of westernization. They selected the NT database as a baseline of biological diversity, subsequently being combined with various studies of interest related to the human microbiota.
This enrichment strategy made it possible to improve classification capacity, compared to the original unenriched database, regarding the various lifestyles and populations studied. The effects of incomplete-taxonomy metagenome-assembled genomes on genome database enrichment were also examined, revealing that, while they are helpful, they should be used with caution depending on the taxonomic level of interest.
In terms of high prevalence, the core analysis revealed a conserved set of bacterial taxa in the healthy human gut microbiota worldwide, despite apparent lifestyle differences. Such taxa show a set of traits, metabolic roles, and ancestral status, making them suitable candidates for a hypothetical phylogenetic core of mutualistic microorganisms co-evolving with the human species.
The most recent strategies in studying the human microbiome are to be presented in our 9th World Congress on Targeting Microbiota- to be held on October 19-21, 2022 in Paris.