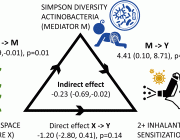
Personalized Whole‐Body Models Integrate Metabolism, Physiology, and the Gut Microbiome
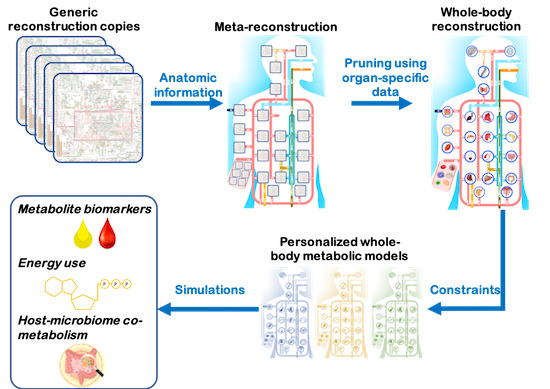 Image from: Mol Syst Biol (2020)16:e8982
Image from: Mol Syst Biol (2020)16:e8982
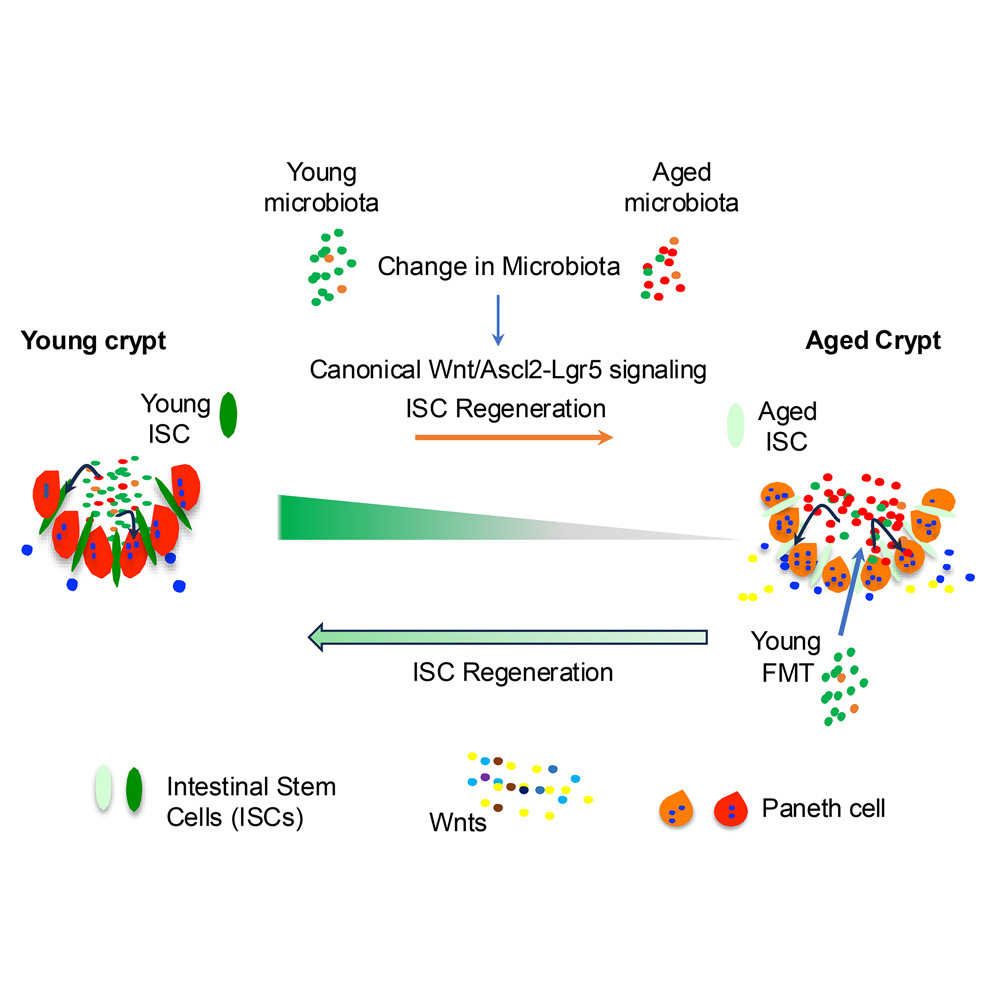
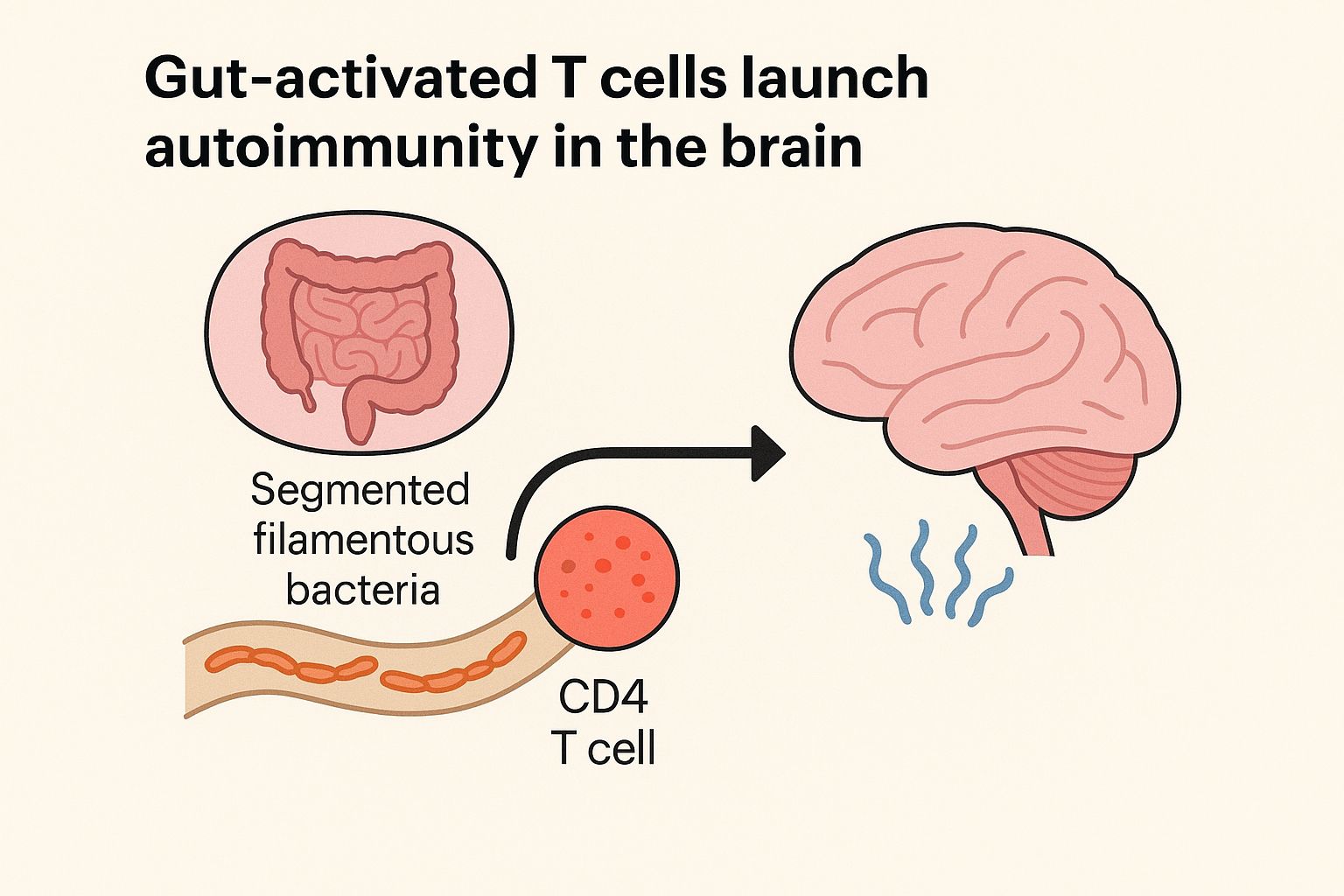
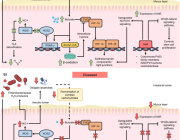
Comprehensive molecular‐level models of human metabolism have been generated on a cellular level. However, models of whole‐body metabolism have not been established as they require new methodological approaches to integrate molecular and physiological data. We developed a new metabolic network reconstruction approach that used organ‐specific information from literature and omics data to generate two sex‐specific whole‐body metabolic (WBM ) reconstructions. These reconstructions capture the metabolism of 26 organs and six blood cell types. Each WBM reconstruction represents whole‐body organ‐resolved metabolism with over 80,000 biochemical reactions in an anatomically and physiologically consistent manner. We parameterized the WBM reconstructions with physiological, dietary, and metabolomic data. The resulting WBM models could recapitulate known inter‐organ metabolic cycles and energy use. We also illustrate that the WBM models can predict known biomarkers of inherited metabolic diseases in different biofluids. Predictions of basal metabolic rates, by WBM models personalized with physiological data, outperformed current phenomenological models. Finally, integrating microbiome data allowed the exploration of host–microbiome co‐metabolism. Overall, the WBM reconstructions, and their derived computational models, represent an important step toward virtual physiological humans.
News Source: www.embopress.org
Journal Reference:
Personalized whole‐body models integrate metabolism, physiology, and the gut microbiome
Ines Thiele, Swagatika Sahoo, Almut Heinken, Johannes Hertel, Laurent Heirendt, Maike K Aurich, Ronan MT Fleming
Mol Syst Biol. (2020) 16: e8982 https://doi.org/10.15252/msb.20198982
Targeting Microbiota 2021 Congress
October 20-22, 2021 - Paris, France & Online
www.microbiota-site.com