Evaluation of the Effect of Storage Methods on Fecal, Saliva, and Skin Microbiome Composition
 Copyright ©️ 2021 Marotz et al.
Copyright ©️ 2021 Marotz et al.The Scientific committee would like to share this excellent article with you.
Abstract: As the number of human microbiome studies expand, it is increasingly important to identify cost-effective, practical preservatives that allow for room temperature sample storage. Here, we reanalyzed 16S rRNA gene amplicon sequencing data from a large sample storage study published in 2016 and performed shotgun metagenomic sequencing on remnant DNA from this experiment. Both results support the initial findings that 95% ethanol, a nontoxic, cost-effective preservative, is effective at preserving samples at room temperature for weeks. We expanded on this analysis by collecting a new set of fecal, saliva, and skin samples to determine the optimal ratio of 95% ethanol to sample. We identified optimal collection protocols for fecal samples (storing a fecal swab in 95% ethanol) and saliva samples (storing unstimulated saliva in 95% ethanol at a ratio of 1:2). Storing skin swabs in 95% ethanol reduced microbial biomass and disrupted community composition, highlighting the difficulties of low biomass sample preservation. The results from this study identify practical solutions for large-scale analyses of fecal and oral microbial communities.
IMPORTANCE Expanding our knowledge of microbial communities across diverse environments includes collecting samples in places far from the laboratory. Identifying cost-effective preservatives that will enable room temperature storage of microbial communities for sequencing analysis is crucial to enabling microbiome analyses across diverse populations. Here, we validate findings that 95% ethanol efficiently preserves microbial composition at room temperature for weeks. We also identified the optimal ratio of 95% ethanol to sample for stool and saliva to preserve both microbial load and composition. These results provide rationale for an accessible, nontoxic, cost-effective solution that will enable crowdsourcing microbiome studies, such as The Microsetta Initiative, and lower the barrier for collecting diverse samples.
Introduction:
Next generation sequencing has enabled an unprecedented view of human-associated microbial communities. This insight has been leveraged to improve understanding of how such communities contribute to human health. Many investigations focus on the gut, using fecal samples as a proxy for the intestinal microbiome, but skin and oral samples are also commonly collected for human microbiome analysis. To comprehensively understand human-associated microbiomes, samples have been acquired from healthy and diseased patients in the clinical setting (1), indigenous peoples in the field (2), and individuals across the globe through community science efforts (3–5). In the clinic, samples are typically frozen immediately at −20°C or below until nucleic acid extraction and sequencing, the gold standard in the microbiome field. However, beyond the clinic, immediate freezing is often not possible, thus leading to the use of preservatives. Importantly, there is currently no standard protocol for microbiome preservation. Identifying a cost-effective, practical solution to microbiome sample preservation could help reduce the barrier to collecting specimens from geographic locations that contain high microbial diversity but are vastly underrepresented (6).
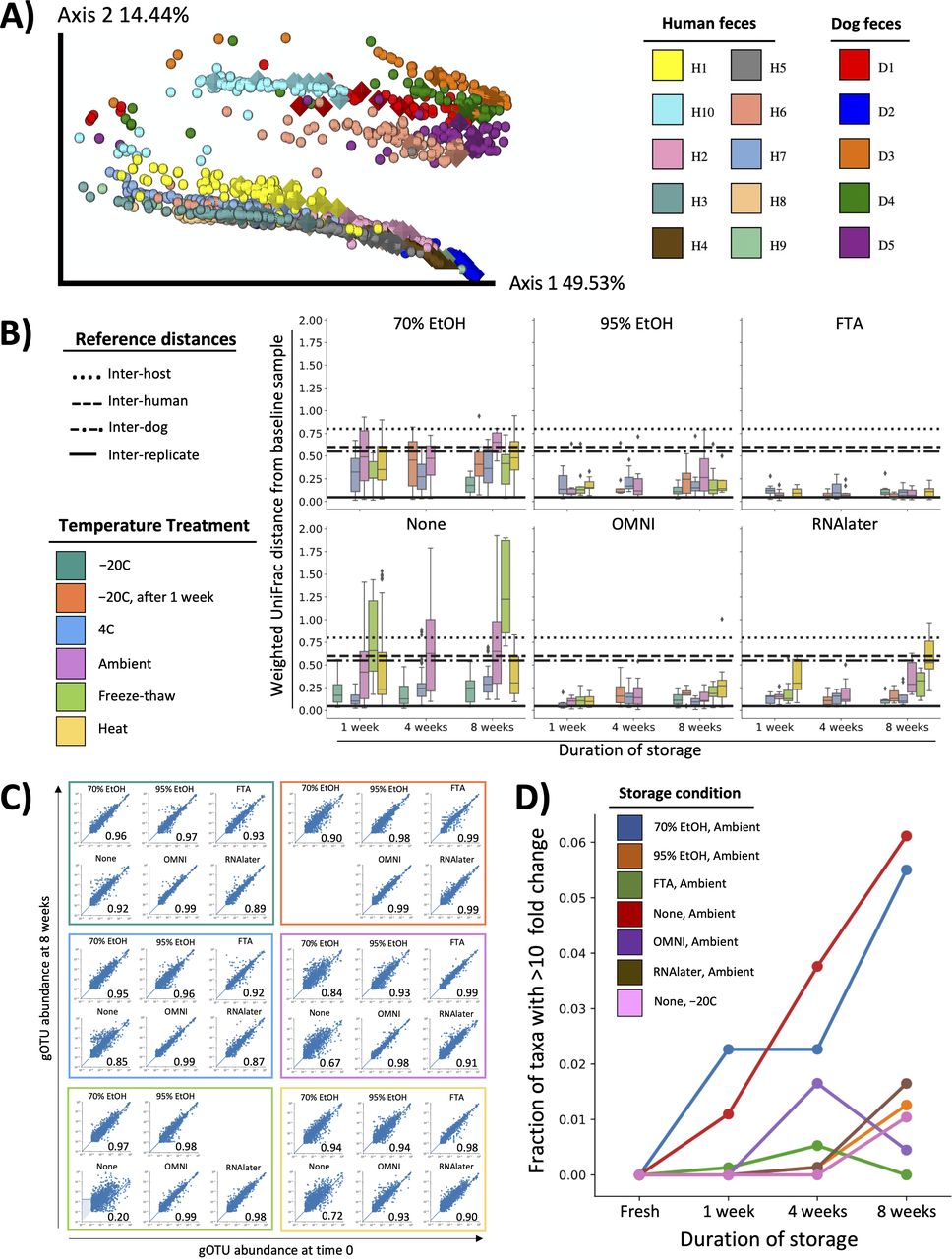
Numerous studies have aimed to understand how fecal storage affects microbiome composition (7–10). In 2016, our group reported one of the largest investigations thus far (11). We performed 16S rRNA gene amplicon sequencing on over 1,200 samples, which included fecal samples from 15 individuals subjected to six preservatives, six storage temperatures, and four time points. Our analysis revealed that composition was well maintained over time and across temperatures when samples were stored in 95% ethanol.
In the last 4 years, microbiome sequencing techniques and the computational tools to analyze next generation sequencing data have advanced considerably. Here, we aimed to improve the understanding of how storage affects microbiome composition using state-of-the-art methods. First, we reanalyzed our original 16S rRNA gene amplicon sequencing data set using a high-resolution exact sequence variant method (12, 13). Next, to further increase resolution (14), we performed shallow shotgun metagenomic sequencing on archived DNA from our original study. Results from these studies confirmed our original conclusion that 95% ethanol is efficacious in preserving fecal microbiome integrity. Finally, we extended our study of 95% ethanol to investigate optimal sample collection protocols for fecal, saliva, and skin samples.
Reference: Clarisse Marotz, Kellen J. Cavagnero, Se Jin Song, Daniel McDonald, Stephen Wandro, Greg Humphrey, MacKenzie Bryant, Gail Ackermann, Edgar Diaz, Rob Knight mSystems Apr 2021, 6 (2) e01329-20; DOI: 10.1128/mSystems.01329-20
Targeting Microbiota 2021 Congress
October 20-22, 2021 - Paris, France & Online
www.microbiota-site.com